-
article · 2026Year9Moon29Day
Flex蛋白质组学工作站的功能与优势
Read More -
article · 2025Year13Moon27Day
Opentrons Flex蛋白质组学工作站:提升实验室效率的秘密武器
Read More -
article · 2025Year42Moon26Day
Flex 工作站在蛋白质组学实验中的最佳实践是什么?
Read More


Authors Jonathan Lin 1 Ryan Sassada 1 , Jessari Red 2 , Homam Jamal 2 , Kinneri Watson 2 1 Zymo Research 2 Optrens Laboratories Inc.
Abstract Microbiology is an emerging field of importance for a broad range of topics in human health. Modern sequencing technology can provide rich and novel data but is limited by DNA extraction methods, which are difficult, time-consuming, and prone to bias, especially in microbial samples. Automated systems can address some of these challenges, but they must be carefully designed and calibrated and paired with appropriate reagents to provide high-quality results. In the current set of tests, Vision Center uses the OT-2 Nucleic Acid Extraction Workstation ZymoBIOMICSTM 96 Magnetic Bead DNA Kit to extract DNA from typical microbial samples. The system and kit were evaluated for yield and purity metrics. Extraction bias and cross-contamination were also assessed separately. Results showed that OT-2 combined using the ZymoBIOMICS kit produced high-yield, high-purity DNA samples without bias and cross-contamination. Several adjustments were identified to further improve performance. In summary, these tests exemplified the accuracy and precision of the OT-2 in implementation across different applications, delivering the best performance of an industry-leading microbiology DNA extraction kit.
Introduction The human microbiota is no longer in the context of biological research. The diverse, large, and highly active microbiota has been in the spotlight and is increasingly valued for its broad role in human health, including roles in infectious diseases, circadian rhythms, and psychological conditions. Automation can support this emerging field of bias-free performance in critical nucleic acid extraction steps. Quality DNA extraction relies on efficient solubilization, which is especially challenging for microbiome samples given the diverse organisms they contain. Some organisms are more resistant to cold and dissolution, while others are more susceptible and dissolve more easily. If a lysis method cannot overcome these differences, hardy species will produce less DNA and more susceptible species will produce more, leading to biased representation in downstream analyses.
Two main lysis methods are commonly used: enzymatic or reagent-based lysis and mechanical lysis. This mechanical lysis kit using bead-based technology in the ZymoBIOMICS 96 Magnetic Bead DNA Kit uses ultra-high-density, Jerk Beads™ to deliver uniform lysis of microorganisms and is compatible with automation because the beads are preloaded in the sample-prepared tubes.
Automation provides further improvements for DNA extraction In addition to the challenges of lysis, manual DNA extraction protocols are time-consuming and prone to variability caused by errors and differences in laboratory technician performance. Automated procedures can eliminate variability and increase throughput, but must be carefully calibrated to ensure that DNA is isolated with good yield and purity.
This study sought to evaluate the performance of the OT-2 nucleic acid extraction automated workstation with the ZymoBIOMICS 96 MagBead DNA microbiology kit. Yield and purity were determined in typical microbiological fecal samples using standard extraction performance specifications. These metrics were compared between automated workstations and automated workstations performed by trained manual process technicians. Automated workstations were also tested for cross-contamination between plate wells.
This test was designed to optimize the protocol of the kit and system. OT-2 avoids extraction bias and delivers samples with yields and purity comparable to manual procedures performed by trained technicians. Yokoi contamination levels are also comparable between the two. Two extraction methods. Additionally, some protocols were adapted using the OT-2 automated workflow to increase throughput and reduce turnaround time.
Results Automated DNA extraction using the OT-2 Workstation and the ZymoBIOMICS MagBead Kit successfully purified DNA without introducing bias. Purification bias can be caused by uneven lysis between microbiota species in a sample. Some species are less prone to solubilization and therefore may be underrepresented in downstream sequencing data. Jerk Beads The technology used in the ZymoBIOMICS kit is a system of mechanical disintegration via a bead mill or bead mill. This technique involves combining the sample with small glass, steel or ceramic beads and forcefully mixing the two. 1 During the mixing process, the beads collide with cells, breaking the cell membrane and cell wall, releasing DNA. Beading reagents use ultra-high-density beads to uniformly dissolve microbial samples in this way, preventing bias.
To evaluate this performance, we tested samples of ZymoBIOMICS microbial community standards (N=8) using the OT-2 Workstation and the ZymoBIOMICS 96 MagBead DNA Extraction Kit. Extracted DNA was analyzed using 16S rRNA gene targeted sequencing with primers targeting the V3-V4 region on the Illinois Sequencing® MiSeq™ instrument. A microbial community standard is a well-characterized reference sample that includes Gram-positive and Gram-negative bacteria as well as yeasts of varying size and cell wall composition. The observed composition produced by the offset fragmentation method does not reflect the theoretical composition of the standard. The sequencing data showed that automatic extraction was performed using the OT-2 nucleic acid extraction workstation and the ZymoBIOMICS 96 MagBead DNA kit, and microbial profiles that met the microbial community standards with high fidelity were observed (Figure 1).
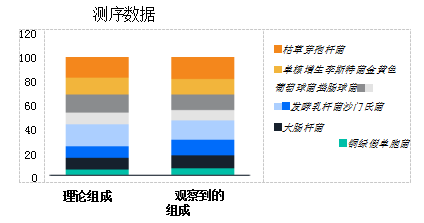
Kit automation provides yield and purity matching the performance of manual extraction of OT-2 nucleic acid extracts Workstation with Enzymatic Microbiology 96 MagBead DNA Kit for DNA extraction from typical microbial fecal samples for DNA yield and purity assessment and with Manual procedures performed by highly trained technicians were compared (N=12). DNA concentration and absorbance ratio (A260/230, A260/280) were measured using Nano Drop2000 UV-Vis spectrophotometer.
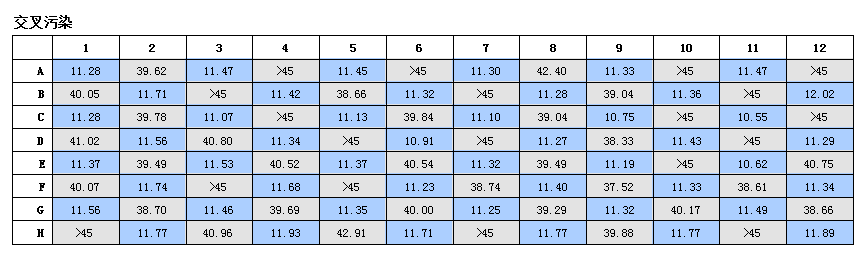
Manual and automated procedures delivered near identical results (Figure 2). The average yield of this procedure for automated and manual operations was 57.88ng/µL and 57.85ng/µL, with average absorbance values of 260/2801.94 and 1.90, and average absorbance values of 260/230 at 1.85 and 1.87 respectively. Automated procedure avoids cross-contamination Cross-contamination between plate wells presents a significant problem with DNA extraction procedures, as contaminated samples can compromise data integrity. However, contamination can be difficult to prevent with manual handling procedures. In contrast, automation can reduce contamination and facilitate audit procedures. If contamination does occur, it will be identified. To evaluate the OT-2 Nucleic Acid Extraction Workstation for cross-contamination, Cryptococcus neoformans and Enzyme Microbiology DNase/Fre-RNase water were added to a 96-well plate and used alternately in a checkerboard pattern. Plates were processed using an automated workstation, and the eluate from each well was subjected to qPCR detection using the Femto bacterial DNA quantification kit on tmCFX96.
The touch real-time PCR detection system detected neonates in any of the water-filled control wells, indicating that no cross-contamination occurred (Figure 3).
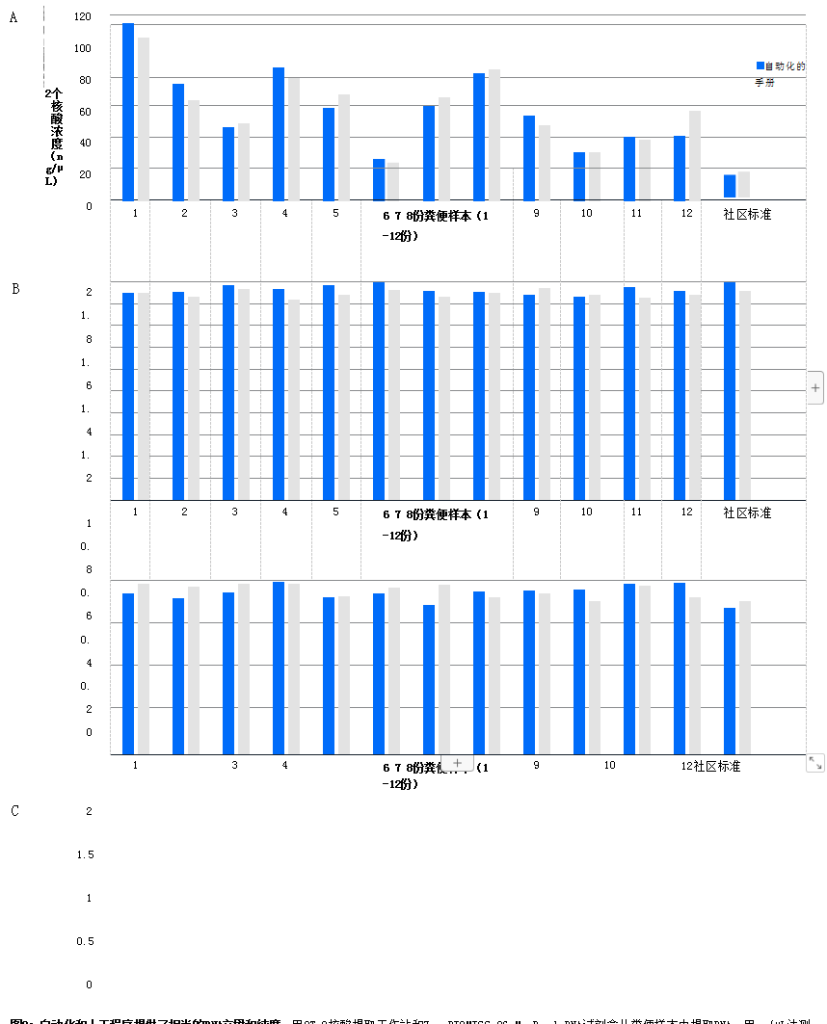
Figure 2: Automated and manual procedures provide comparable DNA yield and purity. DNA was extracted from stool samples using the OT-2 Nucleic Acid Extraction Workstation and the ZymoBIOMICS 96 MagBead DNA Kit. Yield and purity of purified DNA were determined using the ng/µL method, and absorbance values of 260/280 and 260/230 obtained using NanoDrop 2000 UV-Vis spectrophotometry, in terms of yield (A) and purity (B and C ), almost the same results were obtained.
Conclusion The evaluation of the DNA extraction method using the OT-2 nucleic acid extraction workstation paired with the ZymoBIOMICS 96 MagBead DNA kit showed no purification bias. Automated extraction provides yields and purity comparable to manual extraction by trained technicians while avoiding cross-contamination between wells. Additionally, several protocol adjustments were discovered to improve the performance and speed of the extraction workflow.
Use OT-2 nucleic acid extraction workstation and ZymoBIOMICS 96 MagBead DNA kit to extract DNA to obtain high-purity DNA with high yield to purify bias or cross-contamination. excellent
Performance without contamination or purification bias is essential for DNA extraction protocols to be maintained. With the continuous development of microbiological research. These high-performance results demonstrate that OT-2 can provide a reliable purification solution for microbial workflows. The data also demonstrates the flexibility of the OT-2 for use with a toolkit that supports a wide range of applications. OT-2 can provide increased throughput and reduced turnaround time to facilitate more ambitious experiments, saving valuable time while maintaining high standards of quality data.
The experienced service team and strong production support team provide customers with worry-free order services.

简体中文

繁體中文

English

日本語

한국인