-
article · 2026Year31Moon7Day
Flex蛋白质组学工作站如何简化蛋白质样品处理与分析流程?
Read More -
article · 2026Year56Moon6Day
Flex蛋白质组学工作站的自动化功能,如何提升定量精度?
Read More -
article · 2026Year53Moon5Day
Flex蛋白质组学工作站的磁力模块:提升蛋白质纯化效率
Read More


Application Introduction The 2020 COVID-19 pandemic has raged across the world, intensifying human demand for high-throughput RNA detection and gene sequencing for clinical research purposes. Traditional manual operation methods are difficult to improve the efficiency of RNA detection. In this study, we demonstrate an automated SARS-CoV-2 RNA extraction workflow using the Beckman Coulter Life Sciences RNAd-vance Viral Kit (C63510) on an Opentrons OT-2 automated pipetting workstation. We present analytical data showing limits of detection (LoD) for SARS-CoV-2 quantification by qRT-PCR and sequence alignment data that inform automated sequencing samples and library preparation for genetic characterization of mutant strains.
Overview of the RNAdvance Viral Kit and OT-2 Platform The RNAdvance Viral kit efficiently extracts viral RNA from samples by utilizing SPRI paramagnetic bead technology. We used this reagent on an automated Opentrons platform from synthetic nasal matrix (110 mM NaCl, 1% w/v porcine gastric type I mucus (Sigma M2378-100G) and 10 μg/mL w/v human genomic DNA (Coriell NA12878), 90% v/v TE/SNM) samples were extracted with different concentrations of heat-inactivated SARS-CoV-2(ATCC°VR-1986HKTM). Each concentration includes 20 test samples, 2 positive controls, and 2 negative controls.
Workflow throughput Figure 1 shows the OT-2 deck layout for the automated RNAdvance virus extraction workflow. The workflow begins with the addition of Bind (BBB) to the sample lysate and consumes only 10 tips per sample, providing the flexibility to run up to 96 samples in 8 batch sizes. The workflow duration described in the report includes actual operation time, sample lysis time, and optimized sample run time (Figure 2). Using the RNAdvance Viral kit and OT-2 platform, the workflow for processing 96 samples takes a total of 3.5 hours. You can copy the link to the browser to view the specific protocol process: https://protocols.opentrons.com/protocol/bc- rnadvance-viral. You have the flexibility to change method variables based on your needs to get the desired results with your desired batch size, sample input, and elution volume.
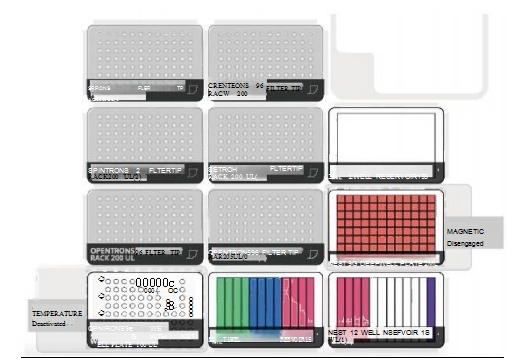
This workflow can be run fully automatically on the OT-2, using 10 tips per sample and starting with the addition of Bind(BBB). The consumables on the experimental deck include: up to 10 tip racks, 1 NEST single-well reservoir, 2 NEST 12-well reservoirs, 1 NEST 0.1mL PCR plate and 1 NEST 2mL deep-well plate. The modules include A magnetic bead purification module and a temperature control module equipped with a P300 multi-channel pipette. All NEST consumables and modules are provided by Opentrons.
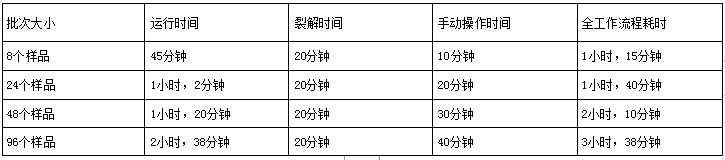
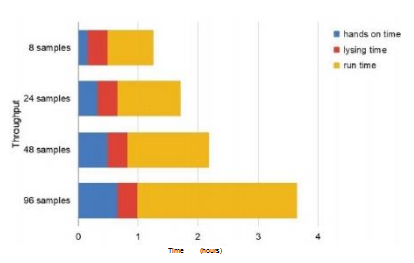
Figure 2: Throughput using the RNAdvance Viral kit on the OT-2 platform. The time required to run the entire workflow consists of 3 parts: operation time, lysis time, and run time. Handling time includes sample and OT-2 preparation. Runtime refers to the time it takes for OT-2 to complete the entire protocol from start to finish. A) It takes 45 minutes to process 8 samples and 2 hours and 38 minutes to process 96 samples. B) Graphical representation of run processing time for different sample numbers.
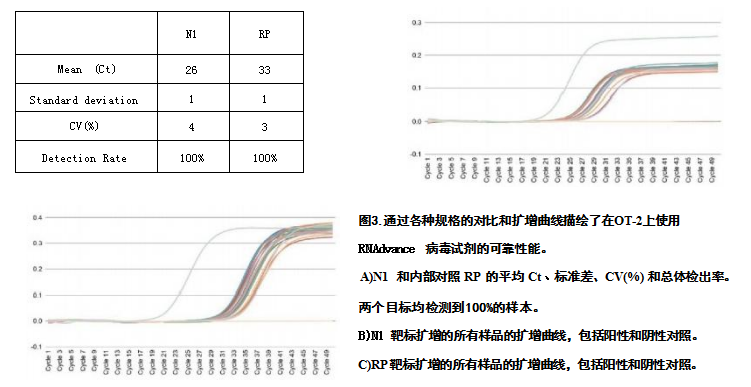
RNAdvance Viral Kit Detection Limit The Limit of Detection (LoD) is a metric used to determine the efficiency of automated RNAdvance Viral extraction on the OT-2 platform. It represents the lowest concentration of viral copy number that can be detected in ≥95% of replicate measurements. We used 2019-nCoV CDC N1 and RP primers and probes (2019-nCoV CDC EUA kit, Integrated DNA Technologies (IDT)) for detection, and the PCR mix and enzyme used Luna° universal probe one-step RT-qPCR kit (New England BioLabs), PCR reactions were performed on a PCRmax ECO48 real-time PCR system. Viral RNA (copies/μL) was detected in SARS-CoV-2 samples (n=20) and positive control samples (n=2), but not in negative control samples (n=2). As summarized in Table 1, all contrast-positive samples within the analysis range were determined to be positive, with the lowest concentration of viral RNA copy number being 1 copy/μL. PCR parameters are as follows.
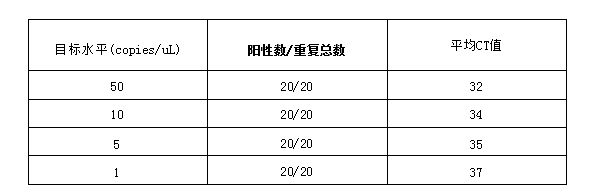
Table 1: Viral sensitivity for detection of SARS-CoV-2 target N1 using the OT-2 platform and RNAdvance Viral Kit. 1 copy/μL is the lowest concentration that can detect ≥95% of the sample and is defined as the limit of detection (LoD)
To study the automated performance of RNAdvance Viral on OT-2, we added heat-inactivated SARS-CoV-2 virus at a concentration of 850 copy/μL into synthetic nasal matrix and extracted RNA. Specification parameters such as mean Ct value, standard deviation, coefficient of variation (CV%), and detection rate were determined for N1 and RP (Figure 3A). In the experimental samples (n=20), positive control samples (n=2) and negative control samples (n=2), the amplification curve of N1 was quantitatively analyzed (Figure 3B), and the amplification curves including the control samples were analyzed quantitatively. RP in the samples was quantified (Figure 3C).
DNA Sequencing and Data Analysis To further understand sequencing performance, we performed N1 amplification using the Swift 2S Turbo DNA Library Prep Kit and Swift Unique Dual Indexing Primer Plates. Detailed data and sequence alignment results for the 850 copy/μl sample can be found in Figure 4 . We observed an average coverage of 455x, and an average of 91% of reads aligned to the reference genome SARS-CoV-2 isolate from Wuhan-Hu-1 (NC_045512.2). The read length matches the N1 amplified region and is approximately 72 bases. According to the Swift 2s Turbo DNA Library Prep Kit manufacturer's instructions, the expected insert size is approximately 200 bp. Sequencing was performed using an llumina MiSeq, and screening data were uploaded to lluminaBaseSpace. Coverage was determined using Bowtie2 in Galaxy and visualized using Integrative Genome Viewer(IGV). The genome of NC_045512.2, SAR2-COV-2, isolated from Wuhan-Hu-1 was the NCBI reference genome used for sequencing. By using the QualiMap-BAMQC tool in Galaxy, information such as alignment, read length, fragment size, and CG content were analyzed.
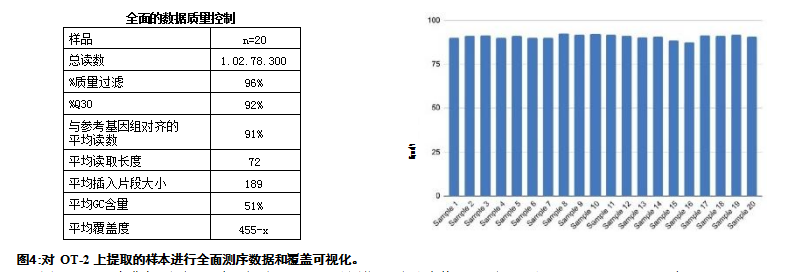
A) Comprehensive data QC table depicting multiple data points including average coverage of 455-x and average reads aligned to the reference genome, 91% for SARS-CoV-2 isolate Wuhan-Hu-1 (NC_045512.2) . The expected insert size according to the manufacturer of Swift 2s Turbo DNA Library Prep is approximately 200bp. B) Comparison of alignment of each sample to the reference genome.
Summary This study provides a stable, cost-effective automated extraction solution using the Beckman Coulter Life Sciences RNAdvance Viral Kit and Opentrons OT-2. This workflow is optimized for SARS-CoV-2 viral RNA PCR detection and measured at a LoD (limit of detection) of 1 copy per microliter. We used high-throughput sequencing (NGS) for testing, achieving an average sample coverage of 455X and an average read alignment to 91% of the SARS-CoV-2 reference genome. This study demonstrates that efficient and cost-effective SARS-CoV-2 viral RNA extraction can be achieved using RNAdvance Viral Reagent and Opentrons OT-2 for sample extraction. At the same time, in terms of sequencing analysis, this solution has high average coverage and high alignment rate, and can provide researchers with important genetic information. This provides an efficient and feasible sample preparation solution for clinical researchers dedicated to studying the SARS-CoV-2 virus.
The experienced service team and strong production support team provide customers with worry-free order services.

简体中文

繁體中文

English

日本語

한국인