Flex应用专题 | 解锁蛋白质谱前处理自动化的无限潜能
Check the Details-
article · 2025Year3Moon9Day
清洗微孔板的具体步骤是什么
Read More -
article · 2025Year56Moon8Day
磁珠分选是什么
Read More -
Press release · 2025Year40Moon8Day
云端相约 | 邀您共同解锁蛋白质谱前处理自动化无限潜能
Read More
AbstractThe demand for PCR is increasing in the life science community. In this application note, we demonstrate that the Opentrons PCR Workstation is able to generate the expected base pair lengths of PCR amplification products compared to popular external third-party thermal cyclers. The results show that both nested PCR and multiplex PCR can produce the expected base pair length of PCR amplification products.
IntroductionThe efficiency and accuracy of complex protocols such as nested PCR and multiplex PCR are difficult to guarantee due to possible contamination, technical errors, and time constraints during sample preparation. Multiplex PCR experiments are used for GMO (genetically modified organism) detection, forensics, food analysis, mutation and polymorphism detection, gene deletion analysis, template quantification, linkage analysis and many more applications. These multiplex reactions are very efficient, producing the products of two or more amplicons in a well.
Nested PCR is another highly specific technique that can be used as a useful diagnostic tool for identifying pathogens in biological samples, such as periodontal pathogens in atherosclerotic plaques, metastatic breast cancer cells, and tuberculosis Unlike standard PCR, which requires only one amplification step for viral pathogens such as mycobacteria and Penicillium marneffei, nested PCR has a two-step amplification process. The PCR amplicon generated in the first PCR amplification step is used as a template for the second amplification step. These multiplex PCR experiments can be tedious and time-consuming depending on the final product. Opentrons PCR workstations automate PCR experiments, requiring only minimal manual preparation and producing consistent results.
Materials and methods
Multiplex PCR Experimental ProcedureA multiplex PCR experiment was performed using genomic DNA isolated from Pseudomonas aeruginosa and Staphylococcus aureus. Based on previous studies, multiple locus region-specific forward and reverse genes for Pseudomonas aeruginosa (lasl, lasR, gyrB) and Staphylococcus aureus (16s rRNA, clfA, mecA, Eubacterial 16S rRNA) were designed. Primers. The expected product sizes for each well are 600, 700, and 222 bp, respectively. On the Opentrons OT-2 platform, 20 μL of multiplex PCR reaction mix was prepared in 0.1 ml 96-well fully skirted NEST plates and amplified using the on-device thermal cycler. The amplified products were analyzed by 2% agarose gel electrophoresis. At the same time, for comparative analysis, we also prepared the same experiment manually and ran it on a third-party manual thermal cycler.
Nested PCR ProtocolNested PCR is a method used to identify pathogens in biological samples isolated from lambda phages. Based on previous studies, forward and reverse multiple-site regions specific for Pseudomonas aeruginosa (lasl, lasR, gyrB) and Staphylococcus aureus (16S rRNA, clfA, mecA, Eubacterial 16S rRNA) were designed. Primer. PCR components were fed to the OT-2 platform and transferred to 0.1 ml 96-well NEST plates. DNA template is added last before mixing and automatic sealing by the Opentrons thermal cycler module. For the second amplification step, repeat this process using a 0.1 ml NEST plate, seal it and place it on a third-party thermal cycler. The amplification product of the first amplification was used as a positive control.
PCR products were analyzed by 2% agarose gel electrophoresis. All experiments were quantified using Qubit BR, and the coefficient of variation was calculated to assess the consistency of amplification.
Experimental resultsUsing the Opentrons thermal cycler module to conduct automated multiplex PCR experiments on the OT-2, the results were comparable to those of third-party thermal cyclers.
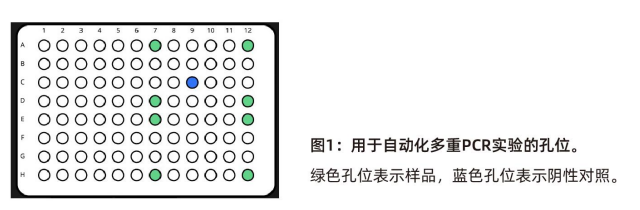
The experimental layout of multiplex PCR adopted the method in the 2018 study by Kim et al. The sample wells are marked in green, and the negative controls are marked in blue (see Figure 1).
To evaluate the high sensitivity and specificity of automated multiplex PCR, PCR product detection was performed on gels. Compared with the PCR products generated by third-party thermal cyclers (Figure 2B), the PCR amplification products generated on the OT-2 thermal cycler not only showed clearer and more obvious bands, but also had the expected length (lasl was 600bp, lasR is 700bp and gyrB is 222bp) (Figure 2A). To further demonstrate the efficiency of OT-2, automated multiplex PCR was performed using specific primers for Staphylococcus aureus (16s rRNA, clfA, mecA, Eubacterial 16s rRNA). The expected base pair lengths were 791bp, 638bp, 499bp and 371bp respectively, and were detected on agarose gel (Figure 2C). Simultaneously, a manual multiplex PCR protocol was performed using the same PCR components, using a third-party thermal cycler (Figure 2). Both automated and manual multiplex PCR on OT-2 showed similar amplification effects (Figure 2A, 2B).
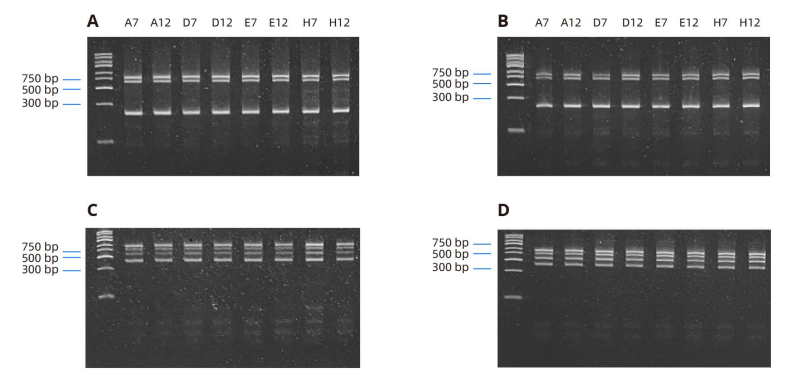
The expected lengths of lasl, lasR, and gyrB targets of 600, 700, and 222 bp were amplified from P. aeruginosa genomic DNA tested on each well. A) Lanes 1-8 show the product after automated loading and thermal cycling on the OT-2. B) Lanes 1-8 show the product after manual addition on a third-party thermal cycler. The expected lengths of 16S rRNA, clfA, mecA, and fungal 16S rRNA targets of 791, 638, 499, and 371 bp were amplified from S. aureus genomic DNA tested at each well. C) Lanes 1-8 show the product after automated loading and thermal cycling on the OT-2. D) Lanes 1-8 show the product after manual addition on a third-party thermal cycler.
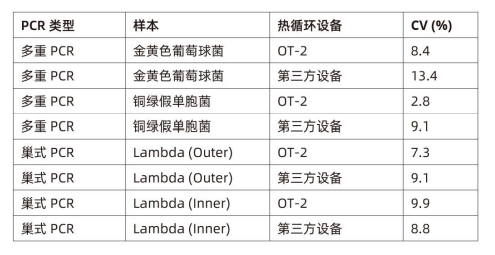
To verify the accuracy of PCR generated on OT-2 and third-party thermal cyclers, the coefficient of variation (CV%) was analyzed, and the results showed that in multiplex PCR and nested PCR experiments, the CV% value of OT-2 was consistent with the The comparison of the three-party thermal cyclers is the same or lower, indicating that the uniformity of amplification is better (see Table 1).
Table 1: Demonstrates highly uniform amplification across different template types and PCR experiments. CV values (coefficient of variation) are calculated after Qubit quantification and are shown for each experiment performed on the OT-2 and a third-party thermal cycler.
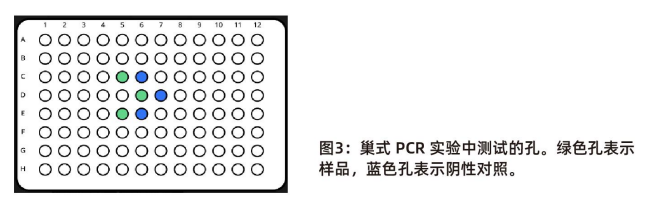
In the layout of nested PCR, the sample wells are marked in green and the negative control wells are marked in blue (Figure 3)
Two sets of primers were designed to detect the 500 bp and 247 bp regions of Lambda template DNA. A 500 bp region was detected in the first PCR run, while a 247 bp region was detected in the second PCR run. Samples were subjected to three replicates of nested PCR, and the amplified products were analyzed at the end of the PCR run.
PCR amplification products generated on the OT-2 and third-party thermal cyclers were analyzed on agarose gels (Figure 4). The expected size of the outer amplified fragment is 500 bp, and the inner amplified fragment is 247 bp. PCR amplification products showed similar yields and coefficients of variation (CV%) (Table 1), and the experimental results illustrate that the output of OT-2 is consistent with manual operation and third-party equipment.

Conclusion
Opentrons PCR Comparison between the workstation and the third-party manual thermal cycler, the alkalinity of the amplified product in PCR It exhibits precise amplification ability in terms of base length and high pipetting accuracy. In addition, theOpentrons PCR workstation demonstrates accurate performance in onewell Amplification capabilities of multiplex PCR and nested PCR experiments.
The experienced service team and strong production support team provide customers with worry-free order services.