-
article · 2025Year5Moon16Day
如何通过 Flex 提高蛋白质组学实验产能?
Read More -
article · 2025Year59Moon16Day
Opentrons Flex 的模块化设计是否有利于实验扩展?
Read More -
article · 2025Year38Moon15Day
Flex 工作站如何帮助实验室实现节能与环保?
Read More


Second-generation sequencing technology is a revolutionary leap in the field of genomics research. Its principles and characteristics have profoundly reshaped our understanding and application of genetic information. The principles and characteristics of Next-generation sequencing (NGS) can be elaborated as follows:
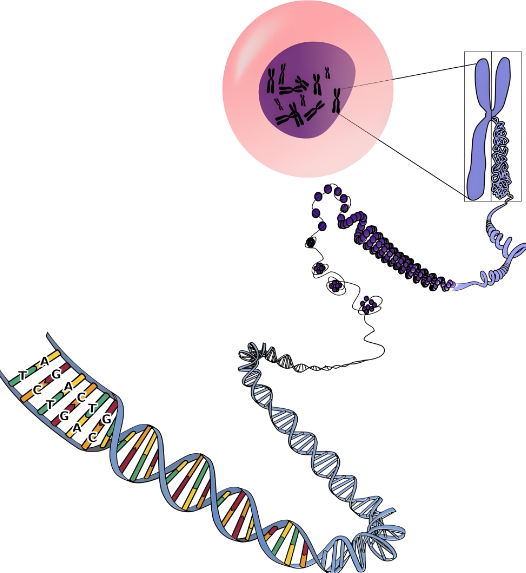
1. Principles of second-generation sequencing
The second generation sequencing technology is mainly based on the principle of sequencing by synthesis (SBS). This sequencing method determines the DNA sequence by capturing special markers carried by newly added bases. Specifically, during the sequencing process, a specially modified DNA polymerase and dNTPs (deoxyribonucleoside triphosphates) with reversible termination fluorescent labels are added to the DNA template strand. Each base of these dNTPs has a different fluorophore attached, and their 3' hydroxyl group is modified so that only one base can be added at a time. When DNA polymerase catalyzes the addition of a dNTP, the fluorophore emits light of a specific color, which is captured and recorded by a detector. Subsequently, the fluorescent group and the group that prevents chain extension are removed through chemical methods to restore the activity of the 3' hydroxyl group and proceed to the next round of sequencing reaction.
2. Characteristics of second-generation sequencing
1. High-throughput: NGS can process millions to billions of DNA fragments simultaneously, greatly improving the throughput and efficiency of sequencing. This makes it possible to sequence a large number of samples in a short time, thus accelerating research in fields such as genomics and transcriptomics.
2. High sensitivity: NGS technology can detect very low concentrations of DNA or RNA molecules and can even distinguish differences between single molecules. This makes it have broad application prospects in fields such as genetic disease diagnosis and early tumor detection.
3. Short read length: Although NGS has high throughput, its single sequencing read length is relatively short (generally tens to hundreds of base pairs). This means that when sequencing a complete genome or transcriptome, DNA or RNA fragments need to be broken into smaller fragments for sequencing and spliced after the sequencing is completed. Therefore, the accuracy and completeness of NGS data largely depend on the splicing algorithm and parameter settings.
4. Low cost: Compared with traditional first-generation sequencing technology (such as Sanger sequencing), NGS greatly reduces sequencing costs. This makes large-scale genome sequencing, transcriptome sequencing and other research more economically feasible.
5. Diversified platforms: There are currently a variety of NGS platforms based on different principles on the market, such as Illumina's HiSeq and MiSeq series, Thermo Fisher Scientific's Ion Torrent series, and Oxford Nanopore Technologies' MinION, etc. Each of these platforms has advantages and disadvantages and is suitable for different research needs and application scenarios.
Second-generation sequencing technology is based on the ingenious principle of sequencing while synthesis, which not only simplifies the sequencing process, but also greatly improves the throughput and efficiency of sequencing, providing unprecedented data support for research in genomics, transcriptomics and other fields.
The characteristics of second-generation sequencing technology further consolidate its core position in the field of life sciences. The continuous optimization of high throughput, high sensitivity, cost-effectiveness ratio, and the continuous emergence of diversified platforms have jointly built a powerful and flexible sequencing ecosystem.
The experienced service team and strong production support team provide customers with worry-free order services.

简体中文

繁體中文

English

日本語

한국인