-
article · 2026Year48Moon30Day
Flex蛋白质组学工作站如何帮助加速蛋白质样品分析?
Read More -
article · 2026Year9Moon29Day
Flex蛋白质组学工作站的功能与优势
Read More -
article · 2025Year13Moon27Day
Opentrons Flex蛋白质组学工作站:提升实验室效率的秘密武器
Read More


Two automated in-house protocols for high-throughput RNA extraction from nasopharyngeal swabs for the detection of SARS-CoV-2 have been evaluated.
141 SARS-CoV-2 positive samples were collected over 10 days. The in-house protocol is based on magnetic bead extraction and is designed for use with the Opentrons OT-2 (OT-2Internal) liquid handling robot or the MagMAX Express-96 system (MMInternal ). Both protocols were tested in parallel with a commercial kit using the MagMAX TM system (MM Kit). Nucleic acid extraction efficiency was calculated based on the SARS-CoV-2 DNA positive control.
No significant differences were found between in-house protocols and commercial kits in detecting positive samples. The MM reagent kit was the most efficient, although the internal MM kit had lower average Ct values than the other two kits. Compared to commercial kits, the in-house protocol saves €350 to €400 per 96 samples extracted.
The described protocol utilizes readily available reagents and open source liquid handling systems and is suitable for SARS-CoV-2 detection in high-throughput facilities.
Citation: Lázaro-Perona F, Rodriguez-Antolín C, Alguacil-Guillén M, Gutiérrez-Arroyo A, Mingorance J, García-Rodriguez J, et al. (2021) Evaluation of two automated low-cost RNA extraction protocols for SARS-CoV-2 detection. PLoS ONE 16(2):e0246302. https://doi.org/10.1371/journal.pone.0246302
Editor: AM Abd El-Aty, Cairo University, Egypt
Received: November 5, 2020; Accepted: January 15, 2021; Published: February 16, 2021 day
Copyright: © 2021 Lázaro-Perona et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Data availability: All relevant data are within the paper and its supporting information files.
Funding: The authors received no specific financial support for this work.
Competing Interests: The authors declare that there are no competing interests.
The SARS-CoV-2 outbreak requires the large-scale use of qPCR testing to detect positive cases and trace contacts to stop community transmission. Before qPCR detection, RNA extraction from clinical samples is usually required [ 1-4 ]. Considering the number of samples tested daily, manual RNA extraction methods are not feasible for most institutions, therefore, automated systems are widely used for this task [5-7]. The disadvantage of an automated system is that it significantly increases the final cost, which may hinder large-scale testing in some areas. Additionally, inventory shortages of extraction reagents caused significant delays in diagnosis due to increased demand.
This article describes two low-cost automated RNA extraction methods. The first method uses the OT-2 system (Opentrons, New York, NY, USA), an open source liquid handling robot capable of automating self-designed protocols; the second method uses MagMAX, a fast and easy-to-use nucleic acid extractor. TM Express-96 system (Thermo Fisher Scientific, Waltham, MA, USA). The latter can extract up to 96 samples in 30 minutes, but requires prior manual distribution of reagents, magnetic beads, and samples in the 96-well plate, which adds 30 minutes. As an alternative, the OT-2internal protocol can process up to 48 samples in 104 minutes, fully automated.
Over ten days, 141 consecutive SARS-CoV-2-positive nasopharyngeal swabs with viral transport medium (Deltalab, Barcelona, Spain) were collected and stored at 4°C. Before processing, 500 μL of virus culture medium and 500 μL of 4M guanidine isothiocyanate (GTC) (Qiagen, Hilden, Germany) were mixed with 5 μg/mL carrier RNA to inactivate the samples, and then the samples were incubated at 80°C. Heat for 2 minutes and vortex briefly to mix.
Two systems were used for automatic extraction of nucleic acids: MagMAX TM Express-96 deep well magnetic particle processor (King Fisher Instrument, Thermo Fisher Scientific, Waltham, MA, USA) and open system OT-2 ( Opentrons, New York, NY, USA) with GEN1 magnetic module (Opentrons, New York, NY, USA) and internal protocol. Both systems used MagMAX™ Express 96 plates and deep well plates (Thermo Fisher Scientific, Waltham, MA, USA).
Three methods were used for nucleic acid extraction: 1) MagMAX and the commercial MagMAX CORE Nucleic Acid Purification Kit (MM Kit) (Thermo Fisher Scientific, Warburg, MA, USA) were used according to the manufacturer's instructions. Elsheim); 2) The OT-2 system uses universal reagents (inside OT-2), such as ethanol (Emsure®, Merck KGaA, Darmstadt, Germany), 2-propanol (Emsure®, Merck KGaA, Darmstadt, Germany), elution buffer (Omega BIO-TEK, Norcross, GA, USA), nuclease-free water (Ambion TM, Thermo Fisher Scientific, MA, USA) Waltham) and magnetic beads (Mag-Bind® TotalPure NGS, Omega Bio-Tek, Norcross, GA, USA); 3) MagMAX uses the same protocol as the commercial kit, but uses reagents from the OT-2 method (MM in-house >). The in-house protocol was a modified version of the procedure described by Hui He et al. [8]. Briefly, inactivated respiratory samples were mixed with isopropyl alcohol in a 1:1 ratio to a final volume of 500 μl (Internal OT-2) and 1000 μL (InternalMM), Add 40 μL of magnetic beads and incubate at room temperature for 5 minutes. Next, use a magnet to pull the beads to the side of the tube and discard the supernatant. The beads were then washed twice with 500 μL of freshly prepared 70% ethanol. After the second wash, discard the 70% ethanol and air-dry the beads at room temperature. Finally, the beads were resuspended in 100 μL elution buffer and separated again with a magnet to recover the eluted viral RNA (Table 1).
Table 1. Steps, reagents, and volumes (μL) used in the three protocols evaluated.
MMKit protocol:
MMinternal protocol:
OT-2internal protocol:
The OT-2 internal protocol is written in Python according to Opentrons instructions. The script is available in a GitHub repository
To validate the performance of the in-house nucleic acid extraction protocol, simulations of standard, low, and very low viral loads were produced using the DNA Positive Control TaqMan 2019-nCoV Control Kit v1 (Thermo Fisher Scientific, Waltham, MA, USA) sample. The concentration of the control kit is 1 x 10 4 copies/μL. Standard viral load samples were prepared by mixing 10 μL of positive control with 490 μL of viral transport medium and 500 μL of GTC. Low and very low viral loads were prepared in the same manner but using tenfold serial dilutions of the positive control. All simulated samples were prepared in triplicate and processed in parallel with InternalOT-2, MMKit andInternalMM, and then used TaqMan 2019- nCoV detection kit v1 for qPCR testing. The final concentrations of positive controls in standard, low and very low viral load mock samples were 1 x 10 6 copies/mL, 1 x 10 5 copies/mL and 1 x 10 4copies/mL. All runs included negative controls.
Calculate nucleic acid extraction efficiency by comparing the Ct value of the positive control in the simulated sample (Ct ss ) with the Ct value of the positive control prepared directly from stock, correcting the amount to match the dilution factor and each protocol The initial sample size used in (Ct pc ) (R = 2 -ΔCt = 2 -(Ctss-Ctpc) ).
Using TaqMan 2019-nCoV detection kit v1 targeting orf1ab, spike (S), nucleocapsid (N) and human RNaseP genes and TaqMan 2019-nCoV control kit v1 as a positive control, Nucleic acid amplification of SARS-CoV-2 viral RNA was performed according to the qPCR conditions recommended by the manufacturer. All qPCR assays were performed in a CFX96 Touch real-time PCR detection system (Bio-Rad, Hercules, CA, USA). To reduce inter-assay variability, extracted nucleic acid samples are automatically dispensed into qPCR test strips using another OT-2 module.
The three alternatives were compared in pairs. McNemar's test was used to compare their performance in assigning samples as positive or negative. Ct values were not normally distributed, so the Wilcoxon signed-rank test was used to compare Ct values for each target. For each target, only samples amplified by these three methods were considered. Use the bootstrap method to calculate mid-position confidence intervals.
All statistical tests were performed using the IBM SPSS Statistics 24.0.0.0 software package (SPSS Inc., Chicago, IL, USA).
The nucleic acid extraction efficiency of the two in-house protocols was compared with the MMkit protocol by extracting mock samples prepared with SARS-CoV-2 positive controls simulating different viral loads. MMKit and OT-2 In-house protocols successfully amplified orf1ab in all simulated samples with standard viral loads , S and N targets. For these samples, the average extraction efficiency across all replicates and genes was 33.9% (SD: 13.8) for the MM kit and 19.6% (SD: 2.2) for the OT-2 in-house protocol. , MMInternal Plan16.0% (SD: 5.03).
In the low viral load simulated samples, the MM kit was unable to amplify the S target in all samples, nor the N target in one of the replicate samples, while the OT-2 internal and MMinternal detected all genes. Finally, the MMkit and OT-2in-house protocols were unable to amplify very low viral load samples, except for one duplicate sample in which the N gene could be amplified by OT-2Internal protocol amplification, but MMInternal could detect the positive control in all samples, one containing three targets and one containing orf1aband N target, the last sample contained theorf1abtarget (S1 Table).
The 141 positive clinical samples collected were simultaneously extracted using MM kit, OT-2internal method and MMinternal method, and analyzed by qPCR Detection of eluted SARS-CoV-2 RNA. Positive/negative results and amplification cycle threshold (Ct) were recorded for each target. Samples were considered positive when at least one of the three target regions was amplified and Ct<40, according to the manufacturer's instructions. Negative controls included in all runs produced negative PCR results in all cases.
Of the 141 samples, 123 tested positive for SARS-CoV-2 by at least one extraction method, while 18 samples tested negative by all three methods. This may be due to degradation of the samples during collection since they had been stored at 4°C for approximately 48 h [9]. By method, 114 samples tested positive using the MM kit, 111 samples tested positive using the in-houseOT-2, and 118 samples tested positive using thein-houseMM . Pairwise comparison found no significant difference in their performance in detecting SARS-CoV-2 (MMKitVs OT-2Internal, P = 0.5465 ; MMKitVs MMInternal, P = 0.3865; OT-2InternalVs MMinternal, P = 0.0961). Eighteen samples tested negative by some of the three methods. These had higher Ct values, with median values of 38.68 and 38.19 for the orf1ab and N targets, respectively (in these samples, the S target was not amplified by any method). Pairwise linear regression and correlation analyzes comparing all methods and targets showed R2 values between 0.85 and 0.95, and Bland-Altman analysis showed consistent agreement across the entire Ct range (S1 Fig ).
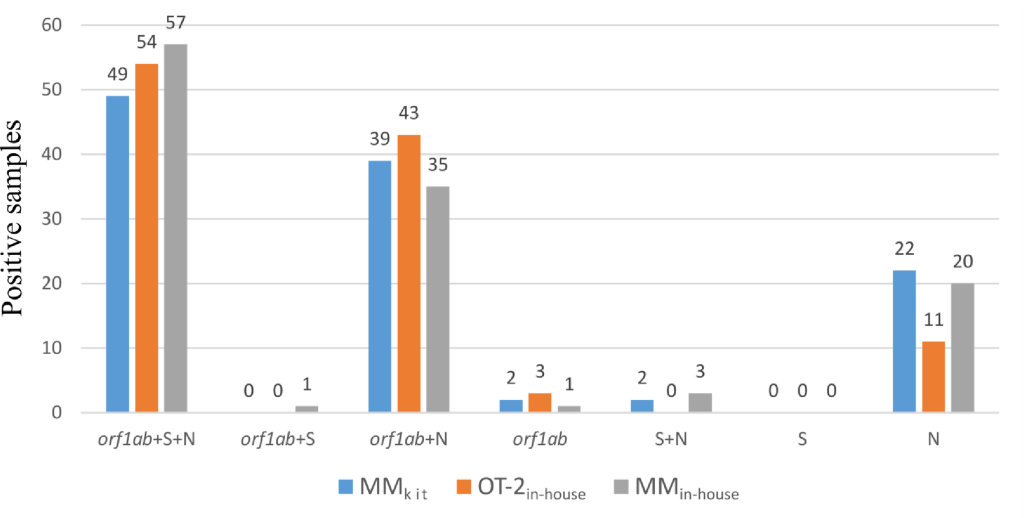
In 43% of the samples extracted using the MM kit, three targets (orf1ab, N and S) were detected, and 43% of the samples extracted using the MM kitInternal It was detectable in 49% of samples extracted with OT-2 and in 49% of samples extracted using in-house MM (Figure 1). orf1aband the N target were detected in 34%, 39%, and 30% of samples, and only the N target was amplified in 19%, 10%, and 17% of samples. Only a very small number of samples were considered positive due to amplification of the orf1ab target alone (6), the orf1ab + S (1) or the N + S (5) target, and no The sample had the S gene as the only positive marker. In fact, the S target in this assay clearly lacks sensitivity and is irrelevant to diagnostic decisions. Two or three targets were detected in 87% (97 samples) of samples using the OT-2internal protocol and two or three targets were detected using the MMinternal protocol. 82% (95 samples), and 79% (90 samples) of samples with two or three targets detected using the MM kit.
There were no significant differences in Cts between the MM kit and the OT-2 in-house protocol in Ct values for orf1ab ( P = 0.437), N ( P = 0.686), and S ( P = 0.794) targets when considering paired samples(Figure 2). With MM kit ( orf1ab ; P < 0.00001 , N ; P < 0.00001 , S ; P = 0.00148 ) and OT - 2Internal Scheme ( orf1ab ; P < 0.00001, Compared with N; P = 0.00008, S; P = 0.00252), the Cts of MMinternal scheme in all targets are indeed significant It's lower.
Figure 2. Box plot showing orf1ab obtained using MMKit, OT-2Internal method, and MMInternal method Distribution of Ct values for em>, S and N targets. The y-axis shows amplification cycles (Ct). Displays the number of data points on each dataset.
Median values for orf1ab targets using MMKit, OT-2Internal Kit, and MMInternal Kit Amplification cycles (CI:95%) were 35.53 (33.82–36.36), 35.58 (34.33–36.21), and 34.8, respectively (33.71–35.29). For the S gene, the median amplification cycles (CI:95%) were 30.16 (28.56–33.08), 31.32 (29.22–32.88), and 31.07 (29.36–32.90); for the N target, the median amplification cycles (CI:95 %) is 34.83 (33.93–35.97), 34.64 (33.42–35.29), and 34.28 (33.52–35.10).
Extraction of nucleic acids from 96 samples using the OT-2 in-house protocol at a total cost of 107 euros (37 euros for reagents and 70 euros for labware), 10 minutes of practical operation and 3 and a half hours of extraction hours (the script processes 48 samples per run, so it must be completed twice). MMinternal costs 66€ (reagents 37€ and labware 29€), MM reagentbox costs 472€ (reagents 443€ and labware 29€) . The hands-on time for both protocols was 40 minutes and the extraction time was 30 minutes. It is worth noting that although the 96-sample OT-2 protocol is 40 € more expensive than the MMin-house protocol, the initial investment required is significantly lower and the OT-2 system (including module and pipette ) is approximately €10,000 and the MagMAX TM system is approximately €50,000.
This article describes two low-cost viral RNA extraction and SARS-CoV-2 detection methods for clinical samples with performance comparable to commercial kits. Although the in-house protocol extracted DNA controls less efficiently than the commercial kit, overall sensitivity was not affected in clinical samples, possibly because the in-house protocol used larger sample input volumes to compensate for the lower efficiency.
Setting up the OT-2 system for RNA extraction requires intensive programming by an inexperienced team, but provides a cost-effective alternative capable of extracting 48 samples in a single run. The scripts used in this work have been uploaded to the open-access software repository GitHub, where many protocols for these and similar applications can be found. This should help other staff avoid or reduce programming steps. This protocol requires little hands-on time and uses readily available reagents. Additionally, the equipment requires lower investment compared to other extraction systems, making it suitable for low- to medium-resource facilities. The MagMAX TM Express-96 is a fast, semi-automatic device capable of extracting 96 samples per run. MMKits provide reagents for inactivation, lysis, washing and elution, and can be used for different matrices and sample types at a competitive cost. The MMinternal protocol, on the other hand, takes advantage of the system's semi-open approach to replace commercial solutions with other chemicals, making them more cost-effective. In both cases, deep well plates, buffers, and samples must be prepared manually, which increases the total extraction time and the risk of operational errors. The main disadvantage of both in-house protocols is that when dealing with viscous samples, mucus, highly viscous polysaccharides, leukocytes, erythrocytes, hemoglobin, proteases, and cellular debris containing large amounts of cellular nucleic acids can hinder RNA extraction or inhibit the PCR reaction [10- 12]. To partially overcome this problem, the sample can be heated and vortexed or centrifuged before extraction, but this increases hands-on time and response time. When using internalMM, the eluted sample may in some cases contain trace amounts of magnetic beads, but this will not affect the performance of the RT-PCR reaction.
Across these three systems, the negative controls were negative in all cases, indicating that there were no cross-contamination issues when processing the original samples in these systems. However, as with non-automated systems, precautions should be taken to separate pre-PCR and post-PCR operations. If the OT-2 module is used to set up a post-PCR reaction (e.g., sequencing), it should not be used to extract nucleic acids from samples unless thoroughly purified.
The main strength of this study is that the methods were tested on real samples in a clinical microbiology laboratory. Comparisons between methods were not systematic, which is an important limitation. In fact, the purpose of design is to optimize the results of each system, so the inputs are different. Systematically exploring input sample sizes as well as other reagent amounts would be useful and informative, but this is beyond the purpose of this study, which was to compare the performance of the system with real samples in a clinical laboratory setting.
In conclusion, the twoin-housenucleic acid extraction methods presented here effectively enable high-throughput diagnosis of SARS-CoV-2 at a fraction of the cost of other commercial kits without loss of sensitivity.
The experienced service team and strong production support team provide customers with worry-free order services.

简体中文

繁體中文

English

日本語

한국인