Flex应用专题 | 解锁蛋白质谱前处理自动化的无限潜能
Check the Details-
article · 2025Year3Moon9Day
清洗微孔板的具体步骤是什么
Read More -
article · 2025Year56Moon8Day
磁珠分选是什么
Read More -
Press release · 2025Year40Moon8Day
云端相约 | 邀您共同解锁蛋白质谱前处理自动化无限潜能
Read More
Abstract Utilizing Opentron's OT-2 and platform, we demonstrate a highly sensitive, cost-effective pipeline for PCR-based detection of SARS-CoV-2.
Key findings. The pipeline can detect trace amounts of virus down to 0.5 copies/μL with ≥96% accuracy on all targets, demonstrating that the sensitivity of OT-2 is comparable to other automated systems. A coronavirus cross-reactivity panel demonstrates the specificity of the SARS-CoV 2 assay compared to SARS, MERS and human coronavirus 229E, NL63 and OC43. .The viral RNA extraction of 96 samples was completed within 3 hours, with an average threshold cycle (Ct) of 36, an average standard deviation of 1.0, and an average coefficient of variation (CV) of 2.8%.
Introduction COVID-19, caused by SARS-CoV-2, has been declared a global pandemic by the World Health Organization. Organization as of March 11, 2020(1). Since then, there has been a strong global focus on developing large-scale, high-throughput testing. While many preventive measures such as social distancing and mask mandates have been implemented in many countries around the world, mass testing has proven to be a powerful option for minimizing the spread of the disease and the threat to the global economy and public health. Countries in the South such as South Korea (2) and Singapore (3) have scaled up extraordinary testing efforts and have become models for containing the disease. A reliable vaccine is still months away (4) and a large number of With testing options showing a high false negative rate (5), increasing the number of reliable testing options is critical.
In this study, we illustrate a simple and reproducible SARS-CoV-2 RT-PCR-based detection strategy using Opentrons OT-2 modules and laboratory software. This automated, friendly workflow is highly responsive, incredibly affordable, and can help immeasurably in increasing testing capabilities worldwide.
Materials and Methods SARSCoV-2 RNA Control 2 (Twist Biosciences) was synthesized and positive samples were synthesized. nasal matrix (SNM) (6), each sample contains 200 μL of lysis buffer and 35 μL of proteinase K (PK). SNM is similar to that outlined in Nuttada et al. (6). (SNM: 110 mM NaCl, 1% w/v mucin from porcine gastric type II (Sigma M2378-100G) and 10 μg/mL w/v human genomic DNA (Coriell NA12878) at 90% v/v TE /SNMEeach samples were lysed by incubating them at room temperature for 15 minutes.
RNA extraction was performed using the Promega® HT Viral TNA Kit and occurs immediately after lysis. After adding the beads and isopropyl alcohol to the sample, pause the mixing plate on the OT-2 for 10 min with the off-deck vortexer. Once mixing is complete, OT-2 is eluted in 65 μL of each sample. . This OT-2 uses a gen2p300 8-channel pipette on the left to mount up to 10 200 μL filter runs, including the GEN2 Magnetic Module and GEN2 Temperature Module (Figure 1). Total run time, including hands-on time and lysis time, varied between 1 and 3.5 hours (Figure 2).
Primers and probes from the U.S. Centers for Disease Control and Prevention (CDC): .2019-nCoV_N1-F (GACCCCAAAATCAGCGAAAT) .2019-nCoV_N1-R (TCTGGTTACTGCCAGTTGAATCTG) .2019-nCoV_N1P (FAM-ACCCCGCATTACGTTTGGTGGACC-BHQ1)•2019-nCoV_N2-F (TTACAAACATTGGCCGCAAA)•2019nCoV_N2-R (GCGCGACATTCCGAAGAA)•2019-nCoV_N2-P (FAMACAATTTGCCCCCAGCGCTTCAGBHQ1)•RP-F (AGATTTGGACCTGCGAGCG)•RP-R (GAGCGGCTGTCTCCACAAGT)•RP-P (FAM-TTCTGACCTGAAGGCTCTGCGCG-BHQ1)
They were ordered from Integrated DNA Technologies (IDT, 2019-nCoV CDC EUA kit). N1, N2 and RNase P were premixed at recommended concentrations 2019 - by CDCnCoV N positive control (GenBank: NC_045512.2) and Hs_RPP30 positive control were both from IDT for SARS-CoV-2 positive control and human RNA positive control respectively . One-Step Starter™ III RT-PCR Kit (Takara Bio Inc.)
Use the Takara or Luna® Universal Probe One-Step RT-qPCR Kit (NEB) at the recommended concentrations tested by Lista, Maria Jose et al. (7). The following procedure performs RT-PCR detection: Reverse transcription results are performed at 55°C for 10 minutes. Initial denaturation was denaturation at 95°C for 1 min, followed by 50 cycles of denaturation at 95°C for 10 sec, followed by annealing at 60°C for 30 sec.
Primers and probes from Charite Berlin/WHO
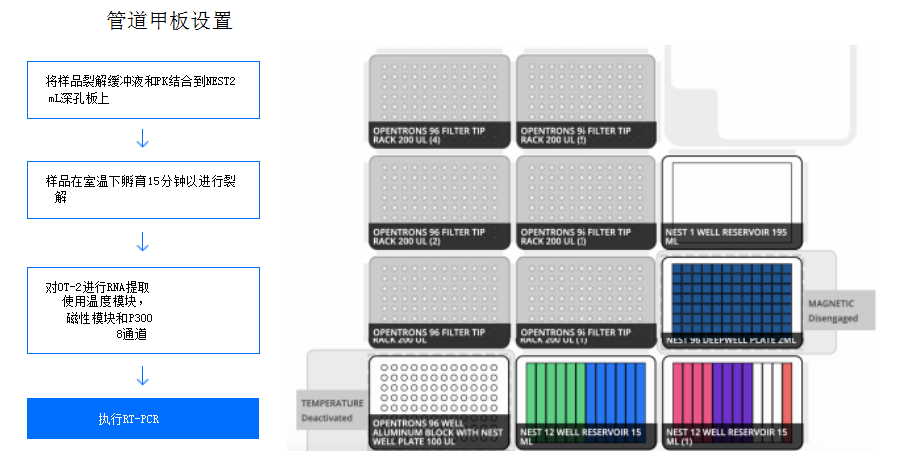
Figure 1: Ease of adoption of SARS-CoV-2 PCR-based detection pipeline A. The pipeline starts with a synthesized sample. After reformatting the samples into 2 mL of NEST 96 deep well plates, the samples were kept at room temperature for 15 min for lysis and then subjected to RNA extraction of OT-2 using the OptoRotor GEN2 Magnetic Module and the GEN2 Temperature Module. Finally, perform RT-PCR on an RT-qPCR machine. This layout on the B.OT-2 includes the GEN2p300 8-channel pipette, GEN2 temperature module, and GEN2 magnetic module. Laboratory software requirements include up to 10 200 µL filter tops, 1 NEST 1-well reservoir 195 mL, one NEST 2 mL deep well plate, two NEST 12-well reservoirs 15 mL, and one NEST 96-well plate 100 µL PCR full skirt. Overall, the total artifact cost per sample was $14.35.
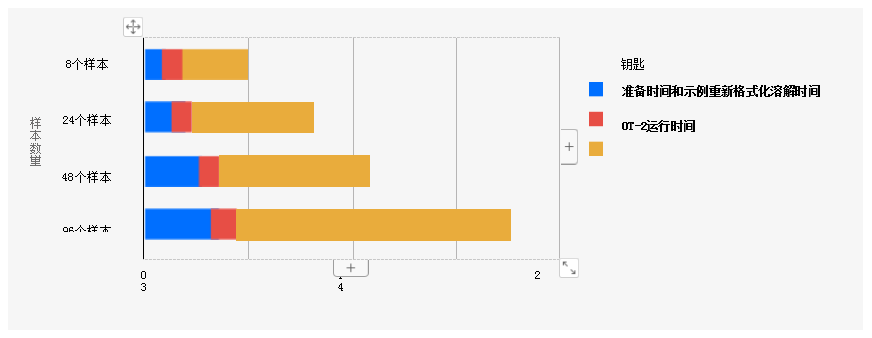
Figure 2: Rapid completion of COVID testing on Opentrons OT-2. Preparation time and sample reformatting time ranges from 10 minutes to 40 minutes, depending on throughput. Lysis time is always 15 minutes. Run times, described as the time it takes for OT-2 to perform a run, varied from 37 minutes for 8 samples to 2 hours to 38 minutes for 96 samples. Overall, 96 samples can be prepared and extracted in approximately 3.5 hours.
Original address:Affordable COVID-19 testing using automation with OT-2
The experienced service team and strong production support team provide customers with worry-free order services.