Flex应用专题 | 解锁蛋白质谱前处理自动化的无限潜能
Check the Details-
article · 2025Year3Moon9Day
清洗微孔板的具体步骤是什么
Read More -
article · 2025Year56Moon8Day
磁珠分选是什么
Read More -
Press release · 2025Year40Moon8Day
云端相约 | 邀您共同解锁蛋白质谱前处理自动化无限潜能
Read More
Summary Three library prep kits—the Illumina® DNA Prep Kit, the KAPA® Ultra Prep Kit, and the NEBNext® Ultra™ II Kit—all automate the rapid and robust preparation of high-quality next-generation sequencing with the Opentrons OT-2 library. 100 ng input libraries were prepared from lambda and human genomic DNA. Similar performance of the libraries, sample variability and yields prepared with the three kits on the OT-2 were observed. Similar ranking metrics such as barcode balance and sequence alignment were also observed for coverage.
Introduction DNA library preparation is a critical step for next generation sequencing (NGS). Automated DNA library preparation provides a simplified method for constructing high-yield sequencing libraries while minimizing hands-on time. Here, we describe the efficiency kit for automating crinoid DNA preparation (Erie, Cat No. 20018705), the KAPA Super Kit (Roche®, Cat No. 7962363001) and the NEBNext Ultra II Kit (New England Biolabs®, Cat No. .E7645L) on the OT-2 robotic liquid handling platform.
Materials and Methods Overview of the Illumina DNA Prep Kit, KAPA Super Prep Kit, NEBNext Ultra II, and Vision Center OT-2 platforms have a patented NGS labeling workflow that utilizes bead-linked transposomes, which is different from the KAPA HyperPrep and NEBNext Ultra II work Process-DNA end repair and A-tail resolution process.
For the Illumina DNA Prep Kit, DNA was labeled and tagged using the Dual Index Adapter on the OT-2. DNA samples (n=8,100 ng) were quantified, normalized to a 4 nM library, pooled into a multiplexed sample, and flow cytometrically run on an Illumina MiSeq on a 2x75 for genomic sequencing. The workflow included the generation of DNA preparation library Lambda (n=24) and human genomic DNA samples (n=8).
For the HyperPrep and NEBNext Ultra II DNA Prep Kits, we constructed DNA libraries of antibody DNA fragments (n=8,100 ng) on OT-2. The barcodes used were from KAPA's unique Dual Index Adapter, and the NEB Multiple Dual Index II set 1 for the NEBNext index. Samples were quantified, normalized, and pooled into a 4 nM library and sequenced on a MiSeq with a 2x75 flow pool. The workflow included fragmented DNA (n=24) and human genomic DNA ( n=8) Generate library.
Sequencing data were demultiplexed by Illumina Basic Space® based on adapter barcode sequences and aligned to the reference genome. Coverage profiles of this genome were analyzed using Geneious Technology® Primary 2021.2.21. Our data show low sample variability and reliable consistency of libraries prepared using the DNA Prep Kit on the OT-2.
Schematic diagram of OT-2 workflow protocol
For Illumina DNA Prep Kits, DNA and IDT® for Illumina DNA/RNA UD Index Set A. .marked on OT-2 (Figure 1). A wash step is necessary to remove residual DNA and adapters. Next, perform PCR amplification using an OT-2 on-deck thermal cycler with programmable lid and block temperatures.
For both HyperPrep and NEBNext Ultra II DNA Prep Kits, NEB fragmentase is used for fragmentation. Human 16 min, human 20 min genomic DNA followed by cleanup with AMPure® , barcode or adapter connection (Figure 1). The barcodes used were from KAPA's Unique Dual Index Adapter (Roche, CatNo. 08861919702). For HyperPrep and NEB Multiplex Dual Index Primer Set 1 (NEB, CatNo. E7335L) NEBNext Ultra II. .
DNA amplification was performed on the OT-2 using an on-deck thermal cycler.
Workflow Layout The layout of the Opentrons OT-2 platform includes modules, laboratory software, and DNA preparation reagents (Figure 2). While the workflow includes automated pipetting on the OT-2, manual intervention is required to move the sample plate between the on-deck thermostat and magnet and to reset the pipette tip rack during this protocol.
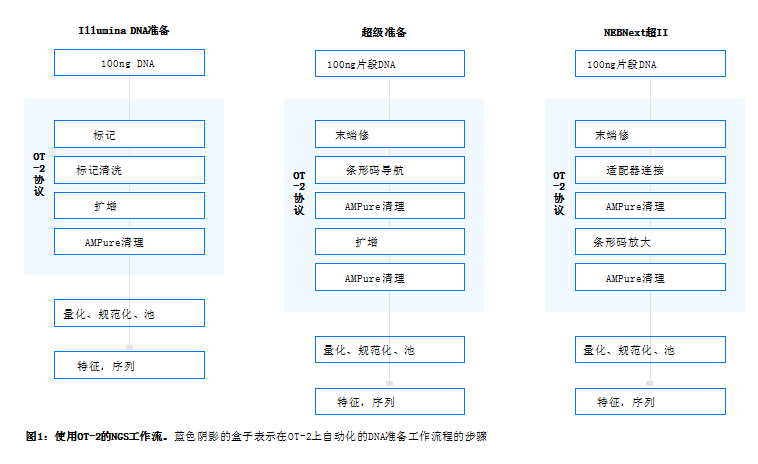
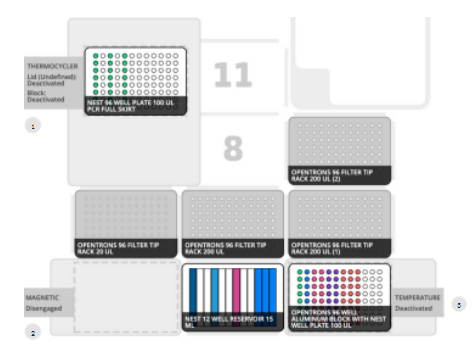
Figure 2: OT-2 deck layout
Modules, laboratory software, and DNA preparation reagents. this
Standardized deck layout shared with HyperPrep and other platforms
NEB Next Ultra II. This workflow automates pipetting on the OT-2 and requires manual intervention to move the sample plate between the on-deck thermostatic pen and (1)
Magnetic block (2), as well as resetting the tip rack during operation. Deck layout includes 1 NEST 12-well reservoir, 1 x 96-well aluminum block, 1 200µl PCR plate on the thermal cycler, 1 NEST 0.1 mL PCR plate on the temperature module (3). Modules include a magnetic module, temperature module, thermal cycler module, and P20 and P300 8-channel pipettes.
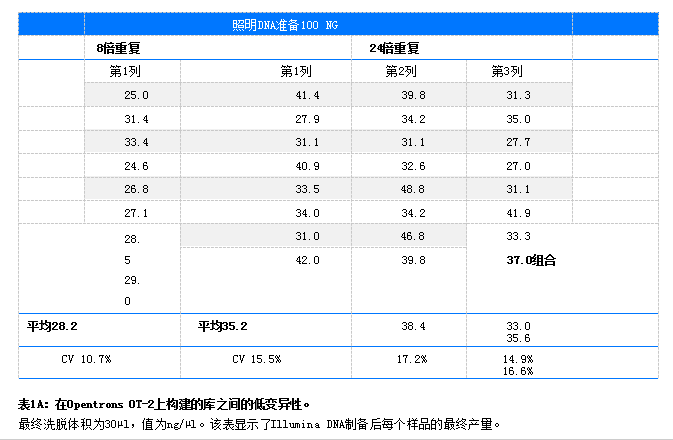
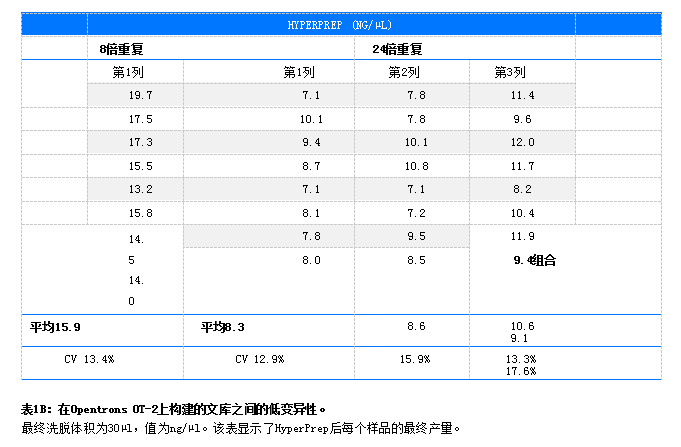
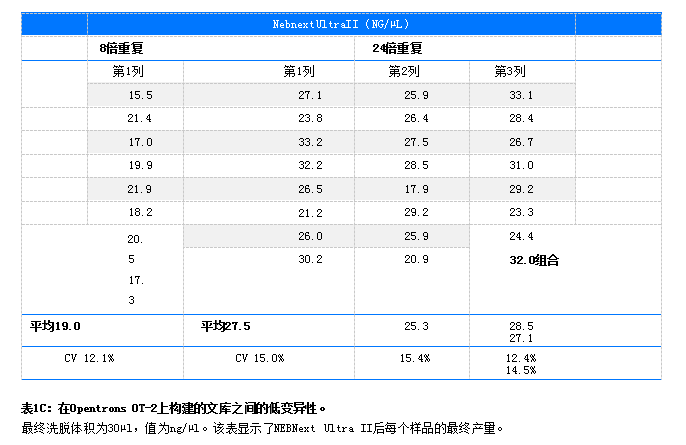
Results DNA library amplification was detected using PicoGreen® in a Qubit® 4. Determine concentration using a fluorometer. CV (coefficient of variation is the standard deviation divided by the mean). The final elution volume for all libraries is 30 µL. For library DNAPrep (n = 8,100 ng) prepared using Illumina the CV was 10.7% and the CV was 16.6% (n = 24,100 ng) (Table 1A). The library was prepared using HyperPrep, with a CV of 13.4% for (n=8100ng) and a CV of 17.6 for (n=24100ng)
. 1B) Libraries prepared using NEBNext Ultra II showed a CV of 12.1% for (=8) and 14.5% for (=24) (Table 1C). A comparison of the yield (ng/µl), CV (%), and fragment size (bp) of lambda and human DNA libraries prepared on OT-2 is shown in Table 2.
Multiplexed DNA prep libraries (n = 8,100) were sequenced on an Illumina MiSeq instrument using a 2x75 flow cell. Second batch of DNA prep libraries (n = 24,100 ng) were sequenced on an OT-2 3 Agilent® 5300 Fragmentation Analyzer calculations are processed in separate columns to account for any column-to-column variability within a batch.
Fragment sizes of DNA prepared libraries were used to determine reliable homogeneity of libraries prepared on OT-2 (Tables 3A-3C). Likewise, consistent sequencing coverage of the 48 kb genome constructed on OT-2 showed consistency in the 8-fold DNA preparation library (Table 4).
Barcode balance is accurate. DNA prep samples showed an average of 11.2%–12.31% for Ilimina DNA Prep, 10.6%–14.1% for KAPA HyperPrep, and 10.5%–14.6% for NEBNext UltraII (Figure 3). Comparing DNA Prep Kits Tip allocation and duration of OT-2 regimen (Table 5).
Conclusion We validated the ImmunoDNA Prep, KAPA Ultra Prep, and NEBNext Ultra II on OT-2 and demonstrated its use in library preparation for next-generation sequencing. We found that DNA Prep samples exhibited low variability in replication and chemistry, even sample barcoding balance, and high sequence alignment coverage, indicating that OT-2 can be used to reliably automate the preparation of high-quality DNA libraries.
Original address:Automatically prepare small genome libraries using DNA preparation kits
The experienced service team and strong production support team provide customers with worry-free order services.